Project P03
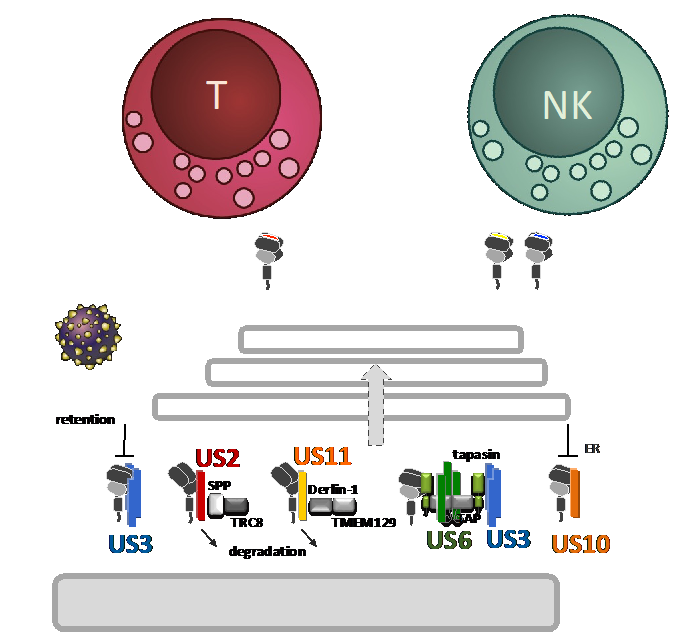
Human cytomegalovirus manipulation of HLA class I by US10
Dr. Anne Halenius
While antigenic peptides presented by HLA class I (HLA-I) on infected cells activate CD8+ T cells, NK cells express both inhibiting and activating receptors engaging HLA-I. This HLA-I-guided multi-axis system challenges strategies of viral HLA-I inhibition and may have promoted the emergence of multifaceted HCMV gene functions that manipulate HLA-I antigen presentation. Interestingly, cytomegaloviruses target antigen presenting molecules by many more strategies than has been found for other herpesviruses, suggesting an exceptionally close co-existence with controlling immune mechanisms. One prominent group of immunoevasins are the type I transmembrane glycoproteins that belong to the US6 gene family. Mechanistically much is known for several proteins in this group, yet, it is not well understood how HCMV orchestrates their function and HLA-I regulation. The mere number of these suggest that they have evolved to control distinct immunological signatures of HLA-I antigen presentation. Underlining this, we recently described that the HCMV glycoprotein US11 preferentially targets HLA-A molecules for degradation, while HLA-B are poor targets for US11. This became strikingly clear when performing HLA-I ligandome analysis of HCMV infected cells. In addition, our recent investigations revealed a close co-evolution of US11 and HLA-A.
Now, we will put a specific focus on US10, a so far insufficiently studied HLA-I immunoevasin. In this project we will perform extensive ligandome analysis (with Z01 and P01) using HCMV deletion mutants and infecting cells derived from various donors, representing various HLA-I haplotypes. We aim at resolving the impact of HCMV encoded immunoevasins on CD8+ T cells and NK cells (with P04 and P07), elucidate further the molecular mechanisms underlying HLA-I manipulation and characterize HCMV derived CD8+ T-cell epitopes.