Project Z01
Analyzing HCMV infection and immune evasion by mass spectrometry
Dr. Andreas Schlosser, Würzburg
Cytomegaloviruses (CMV) express a variety of immunoevasive factors and manipulate MHC peptide presentation in infected cells in various ways to escape elimination by the immune system. Although several targets of CMV immunoevasins have been identified, it remains largely elusive how immunoevasins modulate the immunopeptidome of the infected cells to suppress the presentation of viral antigens. We will combine mass spectrometry-based immunopeptidomics with metabolic pulse-labeling by isotopically labeled amino acids (pSILAC) to accurately quantify the perturbation of the MHC-I antigen processing pathway by HCMV immunoevasins. First, we will study the effect of ectopic expression of individual immunoevasins on the immunopeptidome of fibroblasts. This kind of experiments will not only enable us to study the suppression of antigen presentation by immunoevasins, but also to identify T-cell epitopes associated with impaired peptide processing (TEIPPs) that might be induced by the action of immunoevasins. Next, we will study the modulation of the immunopeptidome by individual immunoevasins in fibroblasts infected with ΔUS2-11 HCMV-infected fibroblasts. This should reveal how individual immunoevasins suppress the presentation of viral T-cell epitopes. Finally, we will perform quantitative immunopeptidome and proteome experiments with wild type and ΔUS2-11 HCMV-infected fibroblasts. These experiments will be complemented by quantitative proteome analyses of different cell types, such as macrophages and endothelial cells, in collaboration with project P01. Together with project P03, we will conduct quantitative immunopeptidome as well as interactome analyses to explore the function of the immunoevasins US10. In addition, we will characterize the ligandomes of various HLA-C alleles and of HLA‑E in HCMV-infected cells. In collaboration with project P02, we will determine the immunopeptidome of MCMV-infected fibroblasts and its manipulation by the viral m04 and MATp1 proteins.
These studies will provide profound insights in the molecular and cellular function of CMV immunoevasins, and potentially in the long run enable the development of new strategies for therapeutic intervention against the immune evasion of HCMV.
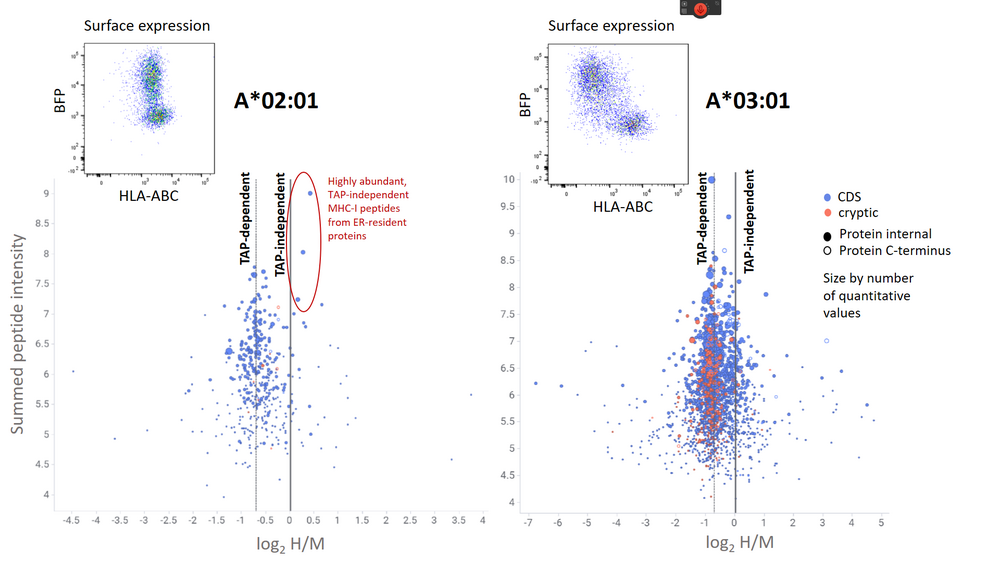
Figure 1: Pulsed SILAC experiment to quantitatively assess the modulation of the immunopeptidome by the HSV‑1 TAP inhibitor ICP47. According to their H/M ratios, most MHC-I peptides from both HLA-A*02:01 and HLA-A*03:01 are TAP-dependent. Conventional MHC-I peptides from coding sequence (CDS) are marked in blue, cryptic MHC-I peptides are marked in red. The quantitative immunopeptidome analysis shows that only a few TAP-independent MHC-I peptides derived from ER-resident proteins rescue HLA-A*02:01 surface expression.